Currently, my research interests focus on most aspects of database, data management, information retrieval and bioinformatics, more specifically, approximate substring search, approximate string mathcing and local alignment algorithms. Besides, I also have done some projects about ad-hoc networks and animal behavior detection when I was an undergraduate.
1.Retrieving Top-K Optimal Similar Substrings under Edit Distance Constraints (ongoing)
The techniques of approximate substring matching can be used in many areas such as information retrieval and computational biology. The general goal of this problem is to match a pattern in a text allowing limited number of errors. However, many answers reported under this definition are overlapping or even contained by each other, thus meaningless. In this paper, we propose the concept of optimal similar substring to avoid the meaningless results. By using this concept we develop a filter algorithm to restrict the candidate set of positions that need to be examined. We then focus on the problem of finding top-k optimal similar substrings under edit distance constraints. We estimate
the distance bounds between the candidates and the given pattern using a novel estimation method. Furthermore, a branch-and-bound algorithm is proposed to retrieve the top-k results. Finally, experiments over real data demonstrate the efficiency of our algorithm. (I will release the source code soon...)
the distance bounds between the candidates and the given pattern using a novel estimation method. Furthermore, a branch-and-bound algorithm is proposed to retrieve the top-k results. Finally, experiments over real data demonstrate the efficiency of our algorithm. (I will release the source code soon...)
2.ALAE: Accelerating Local Alignment with Affine Gap Exactly in Biosequence Databases
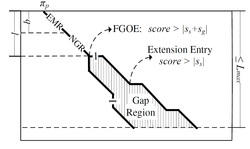
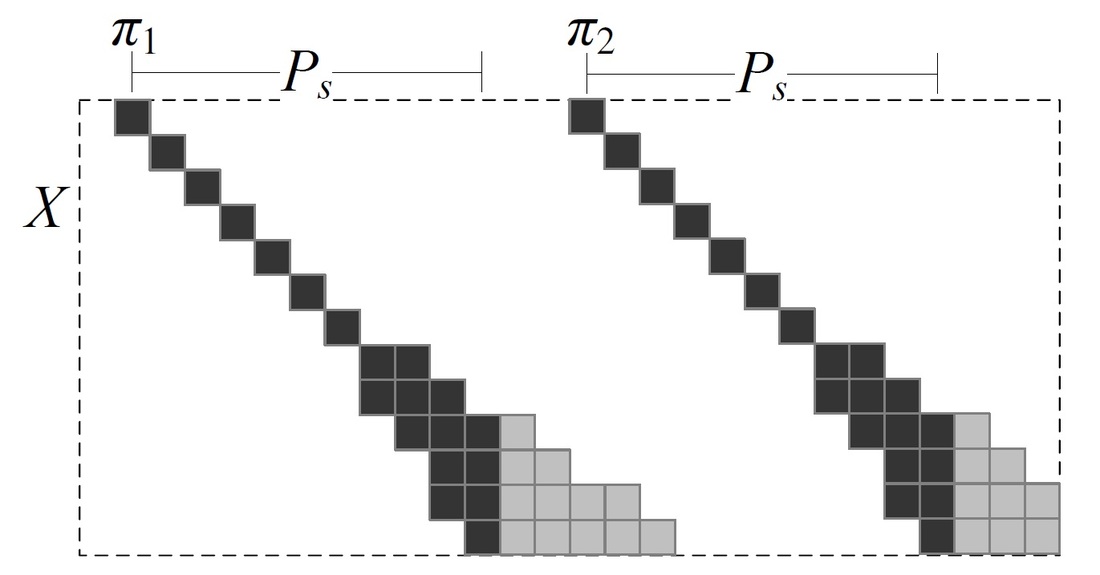
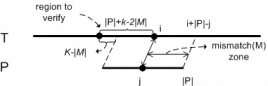
Figure 1. Demonstration of the "Fork Area"
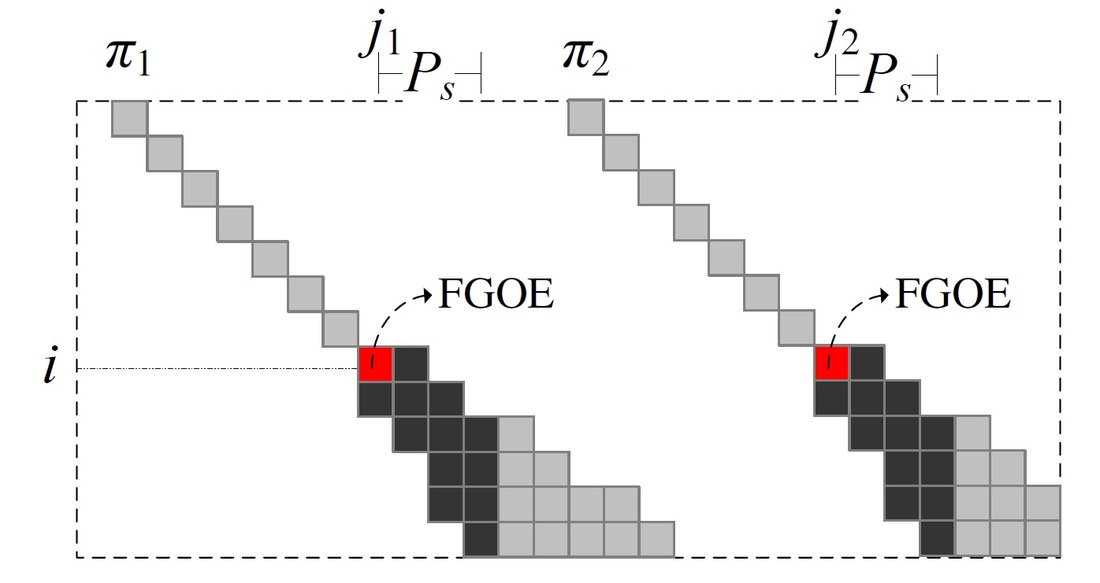
The demand is on finding pairs of similar subsequences with gaps allows, which is called local alignment. The local alignment problem has been widely used in bio-sequence databases. BLAST is a typical software for answering local alignment based on heuristic, but does not guarantee to find all local alignments. Using the Smith-Waterman algorithm, we can find all local alignments between a text T and a query pattern P in O(|T||P|) time, but it is too time consuming especially when the text and the query are long. In this paper, we propose an efficient software called ALAE to speed up the solution of finding all local alignments using dynamic programming and compressed suffix array. ALAE provides a family of filterings (see Figure 1) to prune meaningless calculations and an algorithm to reuse score calculations (see Figure 2 and Figure 3). We demonstrate the efficiency of ALAE using a thorough experimental study on real biosequences. [Source Code is available here]
3.Approximate Substring Query Algorithms Supporting Local Optimal Matching
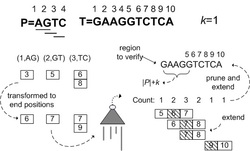
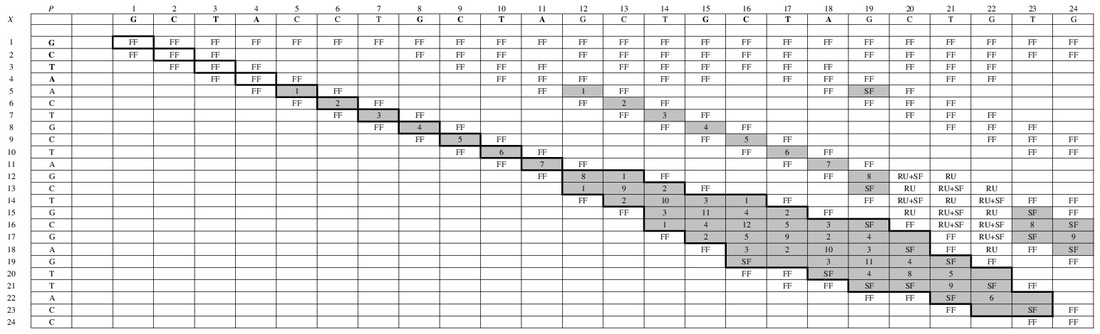
Figure 3. Flow of the algorithm
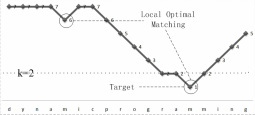
The algorithms of approximate string query which are based on edit distance usually report those strings whose edit distances with the query are not bigger than the given threshold k. However, when talking about approximate substring query, many answers got in this way have overlap, thus meaningless. So we proposed a concept of local optimal matching (see Figure 1) only computing those answers which are both not higher than k and local optimal, thus eliminating the overlapped answers and cutting the time cost. We proposed a definition of approximate substring query supporting local optimal matching along with a query algorithm based on gram index. Moreover, we analyze the generality of the matching process (see Figure 2), and proposed methods of filtering and limiting the boundary based on local optimal matching. Then we offered an algorithm of local optimal approximate substring query with filtering (see Figure 3), so that the efficiency of matching is promoted. [Source Code is available here]
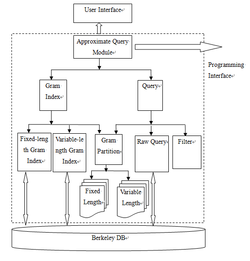
4.An Approximate Querying Approach Based on Berkeley DB
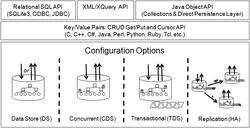
As a very popular open source database, Berkeley DB (see Figure 1) has the same needs. We designed and implemented the approximate query module (see Figure 2) based on Berkeley DB, which is makeup and upgrade of the original query function in Berkeley DB. The system is designed using gram and inverted index technology, containing two major parts which is building index and approximate query. For building index, two ways are provided, namely, fixed length and variable length gram index. For approximate query, this thesis provides a gram segmentation module, a gram index query module and a filter module to achieve the results of user-defined queries.
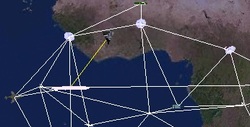
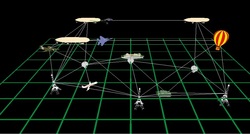
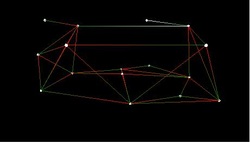
5.Simulation System for Space Ad-hoc Networks
This project was sponsored by the National Undergraduate Innovation Program in China. I participated in the project as team leader, and our team consisted of three undergraduate students. We did some research on simulation of ad-hoc networks and proposed a novel method to demonstrate and evaluate ad-hoc network. We designed and implemented a virtual reality system to simulate space ad-hoc network, which can display the network organization and routing process three dimensionally from multiple views (see Figure 1, Figure 2, Figure 3 and Figure 4). The system makes it intuitive and convenient for the users to analyze the result of the simulation and make better choices.
6.Intelligent Multi-functional Behavior Detection System of Rat

Our project was on exhibition
We implemented this project in cooperation with Shenyang Medical College. Our team consisted of two undergraduate students (see the picture on the left). The project was focused on how to study and learn the behavior of rat effectively and efficiently. We proposed a novel idea of experiment equipment for the above purpose. We also designed and implemented the behavior detection system which can collect the information of rat’s behavior automatically and analyze them easily. Now the system is being used by the Research Group of Disease and Animal Model Learning in Shenyang Medical College for research purpose of animal spatial orientation and cognition.